Article Text
Abstract
Introduction Sepsis management is a challenge for hospitals nationwide, as severe sepsis carries high mortality rates and costs the US healthcare system billions of dollars each year. It has been shown that early intervention for patients with severe sepsis and septic shock is associated with higher rates of survival. The Cape Regional Medical Center (CRMC) aimed to improve sepsis-related patient outcomes through a revised sepsis management approach.
Methods In collaboration with Dascena, CRMC formed a quality improvement team to implement a machine learning-based sepsis prediction algorithm to identify patients with sepsis earlier. Previously, CRMC assessed all patients for sepsis using twice-daily systemic inflammatory response syndrome screenings, but desired improvements. The quality improvement team worked to implement a machine learning-based algorithm, collect and incorporate feedback, and tailor the system to current hospital workflow.
Results Relative to the pre-implementation period, the post-implementation period sepsis-related in-hospital mortality rate decreased by 60.24%, sepsis-related hospital length of stay decreased by 9.55% and sepsis-related 30-day readmission rate decreased by 50.14%.
Conclusion The machine learning-based sepsis prediction algorithm improved patient outcomes at CRMC.
- information technology
- PDSA
- quality improvement
This is an Open Access article distributed in accordance with the Creative Commons Attribution Non Commercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: http://creativecommons.org/licenses/by-nc/4.0/
Statistics from Altmetric.com
Problem
Sepsis, a dysregulated host response to infection, is a serious health concern globally.1 In the USA alone, more than 750 000 individuals are afflicted annually,2 with a cost of over $20 billion per year.3 Many healthcare systems face the challenge of detecting sepsis early with high accuracy. The Cape Regional Medical Center (CRMC), a 242-bed acute care hospital located in Cape May Court House, New Jersey, is one such hospital that has aimed to improve sepsis-related patient outcomes through earlier recognition. This quality improvement initiative details CRMC’s collaboration with Dascena (Hayward, California, USA) to revise their sepsis management system through the implementation of a sepsis forecasting algorithm.
Previously, CRMC had no formal sepsis detection protocol for their emergency department (ED) patients. Patients in all other units were assessed for the presence of two or more systemic inflammatory response syndrome (SIRS) criteria during twice-daily nurse screenings.4 While the SIRS criteria were designed to detect sepsis development, in practice SIRS has demonstrated a low specificity, resulting in a high false alarm rate.5 Additionally, the infrequency of the sepsis screens at CRMC sometimes resulted in delay of treatment during the critical early intervention period.6 In particular, patients who rapidly developed sepsis over a few hours were not always identified in a timely manner with the twice-daily screenings. Consequently, CRMC implemented a sepsis prediction algodiagnostic (algorithmic diagnostic) developed by Dascena to aid in accurate sepsis evaluation.
The sepsis prediction algorithm implemented by CRMC is a machine learning program that provides risk scores that indicate the likelihood of sepsis onset. Through a pre-implementation and post-implementation analysis, we assessed improvements in sepsis-related in-hospital mortality rate, length of stay and 30-day readmission rate with the use of the machine learning algorithm in CRMC’s emergency and hospital patient populations.
Background
Severe sepsis is characterised by organ failure and carries a mortality rate of over 10%, and the escalation to septic shock presents with refractory hypotension and a mortality rate near 40%.7 Despite the high frequency and the poor associated outcomes of sepsis, the heterogeneity of both infection types and host responses makes the early and accurate diagnosis of sepsis difficult. Even consensus definitions of clinical sepsis presentation are difficult to achieve, as indicated by the recent proposed redefinitions of the stages of sepsis (sepsis −3).8 However, it has been shown in several studies that early recognition of sepsis, and compliance with sepsis treatment bundles, can lead to a reduction in patient mortality and length of stay.6 9 Electronic health record (EHR) data are becoming generally more widely available, and represent a rich if complex data source that can be applied to the prediction and detection of sepsis.
Prospective studies of EHR-tool usage in clinical settings have most often been rules-based,10 using predetermined score thresholds to rank patient sepsis risk,11 but demonstrate suboptimal sensitivity and specificity.12 Machine learning algorithms provide potential advantages over rules-based clinical decision support systems, allowing for site-specific adaptability, customisation and predictive capabilities. The machine learning algodiagnostic (MLA) used in this report has been validated in several retrospective studies,13–15 demonstrating a sensitivity and specificity of 0.93 and 0.91, respectively,13 and robustness to missing data.14 Experiments comparing this machine learning algorithm’s predictions with SIRS criteria have also shown that the algorithm’s predictions are more sensitive and specific four hours in advance, as well as at the time of sepsis onset.14 Other prior works applying machine learning algorithms to the EHR in the detection or prediction of sepsis include the retrospective studies of Henry et al 16 and Nachimuthu and Haug,17 and the pilot studies of Sawyer et al.18
Baseline measurement
We collected baseline measurements prior to the revision of CRMC’s sepsis management system. Previously, CRMC used a manual sepsis scoring system, tabulated for all non-ED patients twice per day. Nurses checked each patient every 12 hours, or on identification of a potential source of infection, to determine if they met at least two of the SIRS criteria.19 If a patient met two or more SIRS criteria at the time of assessment, a nurse ordered the nursing sepsis bundle, which comprised a lactate panel, microbiology cultures, a procalcitonin test, a complete blood count panel, a basic metabolic panel, a hepatic function panel and other applicable tests for organ dysfunction assessment. Furthermore, the physician assessed the patient for severe sepsis and accordingly administered all or a portion of the physician sepsis bundle. The physician bundle included the nursing bundle elements together with the following:
fluid resuscitation (normal saline 30 mL/kg intravenously)
broad spectrum antibiotics (meropenem or aztreonam, levofloxacin, vancomycin)
invasive or non-invasive haemodynamic assessment
haemodynamic support (norepinephrine, dopamine, phenylephrine, dobutamine, vasopressin).
Before the implementation of the machine learning algorithm, there was no formalised sepsis screening process for ED patients, but similar interventions were made for patients suspected of or diagnosed with severe sepsis or septic shock.
We extracted retrospective data from 1 November 2016 to 31 January 2017 from each of CRMC’s EHR systems: the Allscripts EHR (San Jose, California, USA) for ED patients and the Cerner Soarian EHR system (Kansas City, Missouri, USA) for intensive care unit (ICU), progressive care unit (PCU) and medical/surgical patients (2East and 4East units). These data represented all measurements recorded for the patient encounters in the baseline time frame.
The primary outcome assessed in this project was the sepsis-related in-hospital mortality rate at CRMC. In addition, the two secondary outcomes measured were average sepsis-related hospital length of stay and the sepsis-related 30-day readmission rate. Patient encounters were included in the sepsis-related outcome metrics if they met two or more SIRS criteria at some point during their stay. Further, only patients over the age of 18 in one of the participating units were included in the assessment. The baseline sepsis-related in-hospital mortality rate was 30/407 (7.37%). The average sepsis-related length of stay during the baseline data collection period was 3.35 days, and the baseline sepsis-related 30-day readmission rate was 188/407 (46.19%).
Design
This project was designed as a prospective quality improvement study. We assessed baseline measurements (pre-implementation) in addition to post-implementation metrics. The first post-implementation phase spanned from 7 February 2017 to 14 April 2017; we additionally analysed post-implementation steady-state measures during the period 20 April 2017 to 20 May 2017, beginning 1 month after the completion of all alert modifications. These data were collected through the EHR systems during the quality improvement project. To oversee implementation of the machine learning algorithm, we formed a quality improvement team overseen by Dr Andrea McCoy, Chief Medical Officer of CRMC. This team comprised clinicians and administrators from CRMC, as well as members of Dascena’s onboarding staff; this team met regularly throughout the post-implementation phase.
The post-implementation phase involved real-time data collection with the use of the MLA. We collected data for all patients in the included units who were over the age of 18. By analysing trends in the collected measurements and their degree of similarity to prior sepsis cases, the algorithm was designed to accurately identify which patients were at the greatest risk of developing severe sepsis. The algorithm used vital sign measurements and, optionally, lab results when present to generate severe sepsis predictions. One of each vital measurement (systolic blood pressure, diastolic blood pressure, heart rate, temperature, respiratory rate and blood oxygen saturation level) was required to generate a prediction score. If a vital measurement was not provided during a given hour, forward-filling imputation (estimation of missing data based on nearby values) was used to gap-fill the missing data.
The algorithm generated a sepsis risk score between 0 and 100 for each patient. Initially, if a patient’s score met or exceeded the threshold of 80, a healthcare provider was called and informed of possible severe sepsis. The CRMC standard of care process, unchanged from the baseline period, was then executed to assess and treat the patient.
Further, to estimate the potential effectiveness of the machine learning algorithm on the CRMC patient population, a side-by-side performance comparison was conducted on an EHR-extracted CRMC data set of 1665 retrospective encounters collected between 1 January 2017 and 30 April 2017. The machine learning algorithm demonstrated significantly higher sensitivity and specificity than the Modified Early Warning Score (MEWS),20 the SIRS criteria, the Sequential (Sepsis-Related) Organ Failure Assessment (SOFA) score21 and the quick SOFA (qSOFA) score22 for a gold standard of severe sepsis, as defined by the patient meeting two or more SIRS criteria and having two or more organ dysfunction lab results (table 1). We additionally assessed the algorithm’s ability to detect sepsis as defined by the Sepsis-3 consensus definition.7 The Sepsis-3 gold standard was defined by suspicion of infection, identified by an abnormal white blood cell count alongside an order of antibiotics within a 24-hour period, and organ dysfunction, identified by an increase in SOFA score of ≥2 points. Sepsis-3 onset was operationalised as the first time both criteria were met within the same hour.
Comparison of AUROC, sensitivity and specificity for the MLA applied to Sepsis-3 and severe sepsis detection, and the SIRS criteria, MEWS score, qSOFA and SOFA scores for severe sepsis detection
We selected severe sepsis as a gold standard for this quality improvement initiative because CRMC uses this definition in diagnosing patients and initiating the sepsis bundle. Despite the 2016 publication of the Sepsis-3 definition, the majority of US hospitals use older definitions of sepsis in diagnosing patients. This is due to several factors, including the relatively recent publication of the Sepsis-3 definition by Singer et al,7 the use of older definitions of severe sepsis and septic shock by the Centers for Medicare and Medicaid Services in their SEP-1 (sepsis core measure bundle) guidelines, and conflicting opinions about the diagnostic utility of the new definition.23 Additionally, the Sepsis-3 definition is quite similar to the chosen gold standard of severe sepsis, as both definitions require evidence of systemic infection alongside organ dysfunction. We have demonstrated success in the past with implementing the Sepsis-3 definition as a gold standard in retrospective studies,13 and the algorithm’s strong performance on CRMC retrospective data under the Sepsis-3 gold standard suggests that the algorithm would likely perform well at CRMC under an updated gold standard, as well.
The MLA was applied to patients throughout the course of their stay (ie, in the ED and/or the hospital). In addition to the use of the algorithm’s prediction scores, CRMC nurses continued tabulation of SIRS criteria every 12 hours for patients in non-ED units, continuing standard procedure as prior to the implementation of the machine learning algorithm; thus, the use of the algorithm did not pose any additional risk to patients. Furthermore, to customise the alerting system to local practice, the quality improvement team regularly incorporated feedback from clinical leadership and end users through the Plan-Do-Study-Act (PDSA) cycles. Modifications resulting from this feedback are provided in the Strategy section. Approval by the institutional review board and informed patient consent were not required in this project as it was a quality improvement initiative at CRMC and did not constitute human subjects research.
Strategy
We conducted three PDSA cycles to evaluate processes and incorporate clinical feedback during this quality improvement initiative.
PDSA cycle 1
The first PDSA cycle focused on the implementation of the machine learning algorithm in the ICU, PCU, 2East and 4East units. Prior to implementation, the quality improvement team held several education sessions to train CRMC clinicians on the proper uses of the sepsis algodiagnostic. Clinicians were instructed to follow all of the aforementioned standard protocols for assessing patients for sepsis. Clinicians were taught how to interpret risk scores and were instructed to continue their twice-daily SIRS screenings in addition to using the algorithm. After implementation, the quality improvement team held regularly scheduled feedback meetings to discuss systemic improvements. Primary areas for improvement concerned the algorithm threshold (initially set at a score of 80) and the reassessment of patients with sepsis.
PDSA cycle 2
The objective of cycle 2 was to best tailor the alerting threshold to workflow and preferences in each implementation unit at CRMC. During cycle 1, clinicians indicated that more patients required bedside assessment, due to the use of the algorithm, than the clinical staff could accommodate. The quality improvement team responded by adjusting the alert threshold to reduce the number of flagged patients, increasing specificity of the alert. During this process, the quality improvement team occasionally manually ran the sepsis algorithm for limited periods to monitor effects of proposed changes. Ultimately, these changes decreased the number of false-positives. Furthermore, per request from end users, the quality improvement team incorporated a 6-hour ‘snooze’ feature to prevent reassessment by the algorithm of any given patient in a 6-hour period. These modifications were well accepted by clinicians at CRMC and enabled focus on caring for the sickest patients without undue time dismissing false alarms. During this cycle, the machine learning algorithm was also implemented into CRMC’s ED. ED staff were educated accordingly on the algodiagnostic’s uses and limitations.
PDSA cycle 3
The final PDSA cycle focused on adjusting the system’s call logic. In a feedback meeting, clinicians addressed the lag time between a prediction score call to a hospitalist and response time to an ED patient. Due to the distance between the ED and other hospital units, it was quicker to direct all ED alerts to a charge nurse or clinical coordinator, rather than to a hospitalist. Accordingly, calls were streamed based on patient location.
Results
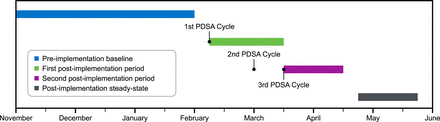
We calculated pre-implementation and post-implementation values for sepsis-related mortality, average length of stay and 30-day readmission rate for a total of 1328 cases, with the pre-implementation period consisting of 407 cases (1 November 2016 to 31 January 2017) and two post-implementation periods consisting of 336 cases (7 February to 14 March 2017) and 381 cases (15 March 2017 to 14 April 2017), as well as 204 cases in the post-implementation steady-state period (20 April to 20 May 2017). Patient demographic data for each period are presented in table 2. The first PDSA cycle was completed in early February, before the start of the first post-implementation period. The second PDSA cycle was completed in early March and the third in mid-March, both before the commencement of the second post-implementation period. The post-implementation steady-state period began 1 month after the completion of all PDSA cycles. The full timeline for the quality improvement initiative is summarised in figure 1. No patients were excluded in the analysis if they were over the age of 18, met two or more SIRS criteria at some point during their stay, and were admitted to one of the implementation units at some point during the analysis time frame.
Demographic characteristics of patients involved in the quality improvement initiative, based on data abstracted from the electronic health record
Timeline of patient outcome measurement collection periods and Plan-Do-Study-Act (PDSA) cycles for the study.
In addition to the qualitative feedback collected during this initiative, we monitored and evaluated patient outcomes. The primary observed outcome of sepsis-related in-hospital mortality rate decreased during the quality improvement initiative. At the pre-implementation baseline, 30 of 407 patients (7.37%) were deceased in-hospital; however, after implementation of the machine learning algorithm and completion of all three PDSA cycles, this proportion decreased to 12 of 381 (3.15%), a 57.3% reduction. In the post-implementation steady-state period (ie, 1 month after completion of all PDSA cycle), this proportion remained steady at 6 of 204 (2.94%), a 74.94% reduction relative to the pre-implementation baseline. Average mortality over the entire post-implementation period was 2.93%, for a 60.24% (p<0.01) average reduction.
Secondary outcomes included sepsis-related hospital length of stay and sepsis-related 30-day readmission rate. Average sepsis-related hospital length of stay improved from 3.35 days to 3.19 days to 2.94 days, a 4.8% and 12.1% reduction, respectively, relative to the pre-implementation baseline, and remained consistent at 2.92 days in the post-implementation steady-state period. The average length of stay over the total post-implementation period was 3.03 days, a 9.55% decrease (p=0.077). This also provided a potential financial impact—an average length of stay reduction of 0.43 days (the difference between the pre-implementation baseline and post-implementation steady-state average length of stay) at an average cost of care per day of $2311 for 300 cases per month (approximate average number of cases per month across post-implementation periods) translates to approximately $3.6 million of cost savings per year to the CRMC healthcare system. Additionally, in the pre-implementation period, 188 of 407 (46.19%) patients with sepsis were readmitted to the hospital within 30 days of initial discharge; however, after the completion of the three PDSA cycles, this proportion decreased to 96 of 381 (25.2%), a 45.4% reduction. In the post-implementation steady-state period, this proportion further decreased to 16 of 204 (7.84%). The average 30-day readmission rate over all post-implementation months was 23.03%, a 50.14% reduction (p<0.01). These results are summarised in table 3 and figure 2.
Comparison of sepsis-related in-hospital mortality rate, hospital length of stay and 30-day readmission rate before and after implementation of the machine learning algorithm. The first, second and steady-state periods all occurred post-implementation.
Outcomes in (A) sepsis-related mortality, (B) sepsis-related length of stay and (C) sepsis-related 30-day readmissions before implementation of the machine learning algorithm and in each post-implementation period, including the April–May post-implementation steady state.
CRMC also experienced an improvement in the 3-hour severe sepsis SEP-1 bundle compliance.24 The average annual 2016 SEP-1 bundle compliance rate at CRMC was 49%; however, this rate increased to 72.7% following the use of the MLA. Early intervention has been shown to reduce sepsis-related mortality, and strong bundle compliance rates align closely with the measured patient outcomes in this initiative.6
Lessons and limitations
This quality improvement initiative was implemented in a community hospital, which typically has cost and mortality outcomes distinct from large academic medical centres.25 26 Often it can be difficult to effectively translate advances in clinical research to a community hospital setting.27 In addition to the successes of this project, there are several limitations related to the generalisability and design of this quality improvement initiative. Because this work was conducted in a 242-bed community hospital in southern New Jersey, it does not provide evidence that the same improvements would necessarily be achieved at a larger medical system or in a different geographical region. Furthermore, this initiative did not have a randomised design. Thus, we cannot draw conclusions about cause-and-effect relationships between the updated sepsis management system and readmissions, length of stay or mortality. Rather, we have documented the steps to an effective implementation of the machine learning algorithm, which was likely associated with such improvements.
Further, it is important to note that we did not analyse this patient population based on International Classification of Diseases (ICD) coding for sepsis, severe sepsis or septic shock. Rather, we based our analysis on meeting two or more SIRS criteria during the patient stay. We chose this method both because ICD code documentation is known to be an inaccurate indicator of clinical diagnosis28 and because of the predictive design of the machine learning algorithm. The algorithm may have enabled earlier intervention and averted development of sepsis or escalation to severe sepsis or septic shock. In addition, due to this patient selection, the absolute values of the outcome metrics (ie, mortality rate, length of stay and readmission rate) may not be directly comparable to other published studies and benchmarks, which often segment populations based on ICD coding; thus, we have chosen to focus on the relative, pre-implementationand post-implementation differences in this report.
Possible confounding factors relate to the personnel involved in sepsis care and management, the frequency of SIRS criteria tabulation, and variation in the baseline period and quality improvement period. Variations of clinical staff in the five units during the pre-implementation and post-implementation time periods may have influenced outcomes such as bundle compliance and sepsis-related length of stay through the level of care individual clinicians delivered. Increased bundle compliance during the post-implementation period, due to either increased awareness of sepsis caused by the algorithm or to other confounding factors, may have resulted in the improved outcomes noted in this study. However, increased bundle compliance may also represent evidence that the alerting system is appropriately drawing attention to patients with sepsis and prompting timely care; the cause of increased compliance cannot be determined by this quality improvement initiative alone. Furthermore, the use of the machine learning algorithm may have increased clinician awareness and unknowingly prompted clinicians to check SIRS criteria more frequently than every 12 hours. Finally, differences in contextual elements of the time periods could have impacted the results. If the pre-implementation period, for example, was a particularly busy time period for the hospital relative to available staff, a greater number of sepsis cases could have been overlooked.
Conclusion
In this initiative, we implemented a machine learning algorithm to improve sepsis-related patient outcomes at the CRMC. With adaptation to clinical feedback, the use of a machine learning sepsis algorithm was associated with improving sepsis management and patient outcomes at CRMC, and by extension the financial burden to the healthcare system. We will continue to monitor the algorithm’s impact on patient outcomes at CRMC and in other care settings to assess the generalisability and acceptance of this system in different hospital environments.
Acknowledgments
We gratefully acknowledge Anna Lynn-Palevsky, Jana Hoffman and Emily Huynh for assistance with editing, and we acknowledge the Dascena algorithm development staff for their work in designing the machine learning algorithm.
References
Footnotes
Contributors Both authors listed in this manuscript contributed to the design and implementation of this quality improvement initiative. AMC oversaw on-site implementation of the machine learning algorithm, patient safety measures, workflow integration and facilitation of clinician feedback. RD contributed to the implementation of the machine learning algorithm, as well as updates made to the algorithm during the quality improvement initiative. RD and AMC contributed to data collection and analysis. Both authors assisted in drafting and editing of the manuscript. Both authors have had the opportunity to draft and revise this manuscript and have approved it in this final form.
Competing interests RD is an employee of Dascena.
Provenance and peer review Not commissioned; externally peer reviewed.
Data sharing statement No data obtained from Cape Regional Medical Center in this study can be shared or made available for open access.